Address : 101Longmian Avenue, Jiangning District, Nanjing 211166, P.R. China
Email : wangmjseven@126.com
HumanTestisDB is a comprehensive database for human testis, it consists of 46 single-cell sequencing datasets from human testis, covering ages from 6 weeks post-fertilization to 49 years old. After uniform analysis, a total of 131,113 cells were retained and classified into 38 different cell types. HumanTestisDB offers six cell sets, each focusing on critical biological events during spermatogenesis and providing in-depth cell type annotations. The 'TestisAtlas' module allows for detailed inquiries into each cell set, including cell features, gene expression patterns, differentially expressed genes for each cell type, and dynamically expressed genes during cell development. HumanTestisDB also elucidates cell-cell interactions within the testicular microenvironment across eight developmental stages, detailing the signaling pathways, receptor-ligand pairs, and the cell types involved in these interactions, which is available in the 'CellInteraction' module. In summary, HumanTestisDB provides invaluable insights into testicular transcriptomics and interactions, and serves as a vital tool for further research in reproductive biology.
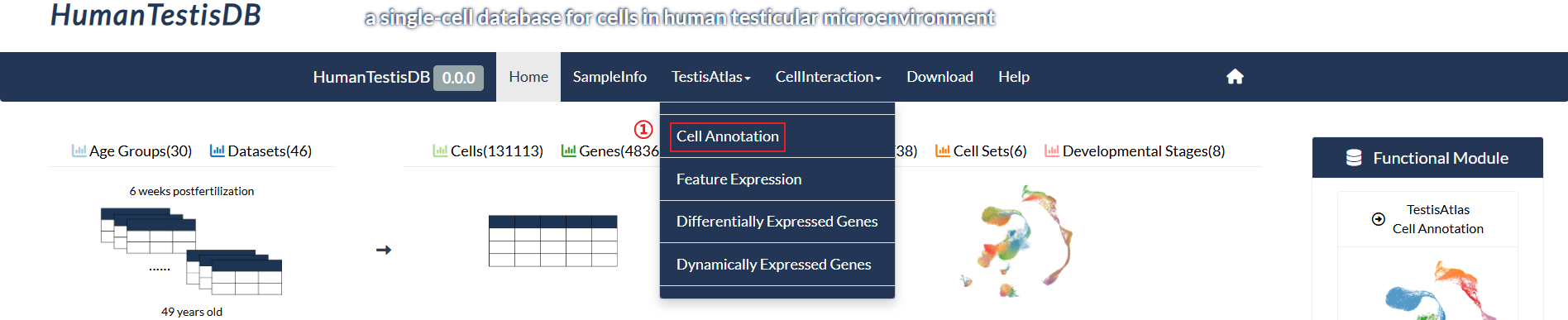
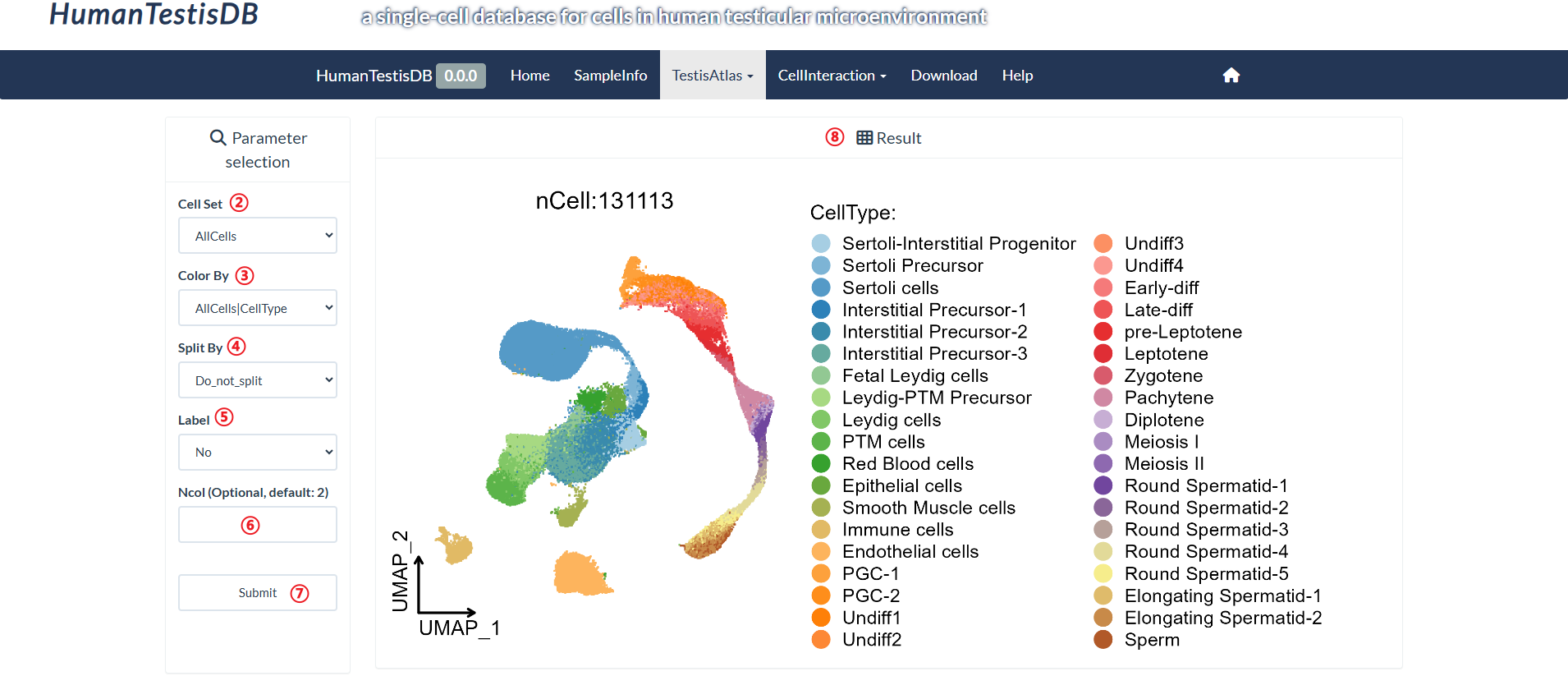
How to explore "Cell Annotation" page?
Users can query UMAP(Uniform Manifold Approximation and Projection) plots of 6 cell sets separately, where each point is a cell. Cells are colored by their identity class (can be changed with the "Color By" parameter).
step1 : Click "Cell Annotation" in the pull-down menu of "TestisAtlas".
step2 : Select the cell set to query in "Cell Set".
step3 : Select a variable to group cells in "Color by".
step4 : Select a variable to split cells in "Split By".
step5 : Select whether to label cell groups in "Label".
step6 : Input number of columns for display in "Ncol".
step7 : Click "Submit" to query result.
step8 : The result is on the right panel.
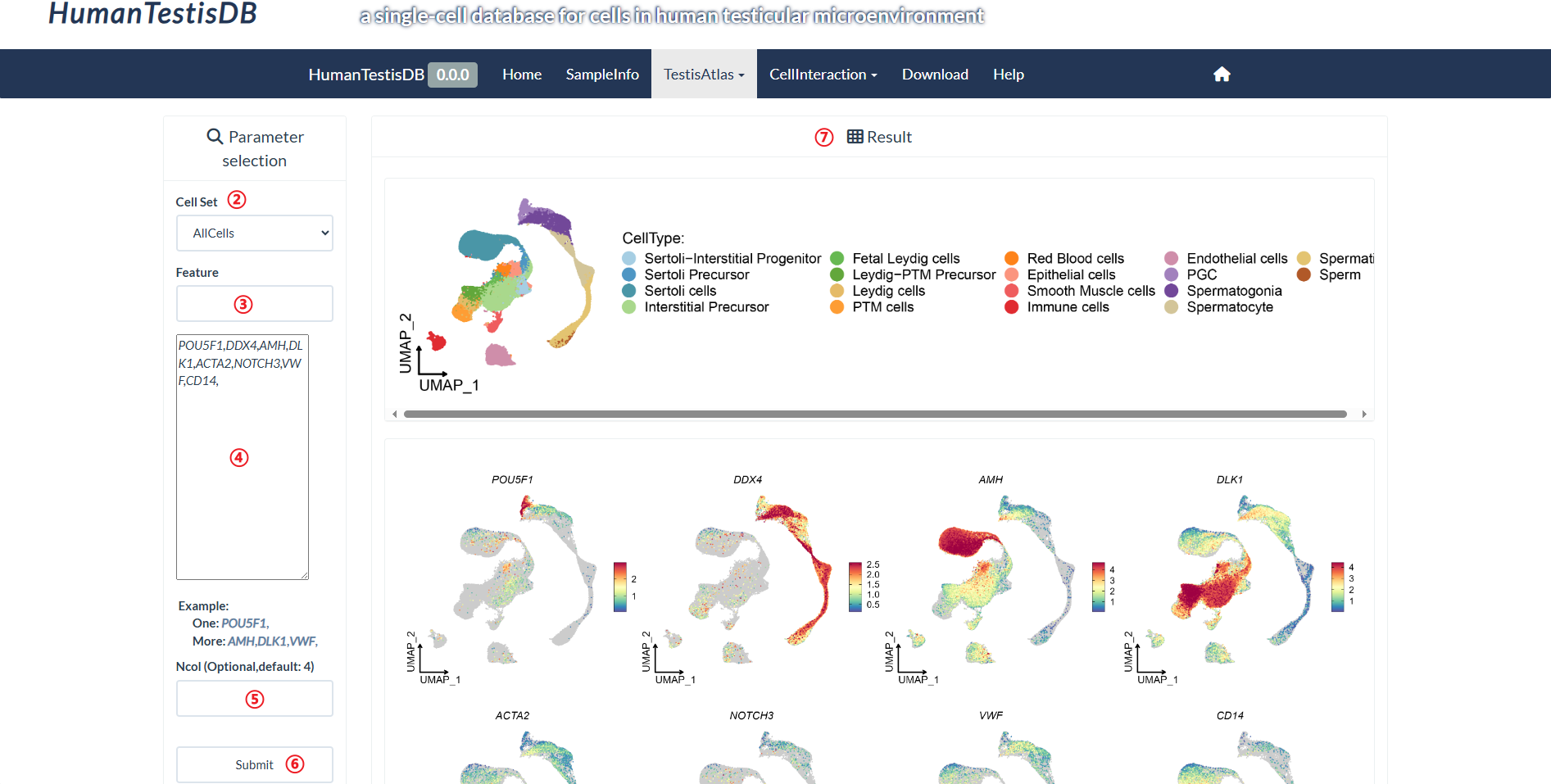
How to explore "Feature Expression" page?
Users can query UMAP(Uniform Manifold Approximation and Projection) plots of 6 cell sets separately, where each point is a cell. Cells are colored according to a 'feature' (i.e. gene expression, the percentage of transcripts that map to mitochondrial genes, number of genes detected, etc.). Note: If the value of "Feature" is empty, the right panel will display the results generated using the default value of "log10_nFeature_RNA".
step1 : Click "Feature Expression" in the pull-down menu of "TestisAtlas".
step2 : Select the cell set to query in "Cell Set".
step3-step4 : Select/Input one or more features (using commas as the separator) in "Feature".
Provide two ways to select genes. The first way is to input at least two characters in the region marked as 3, which will provide features containing those characters for selection. The selected features will be displayed in the region marked as 4. The second way is to directly input the features to be queried in the region marked as 4, using commas as separators.
step5 : Input number of columns for display in "Ncol".
step6 : Click "Submit" to query result.
step7 : The result is on the right panel.
How to explore "Differentially Expressed Genes" page?
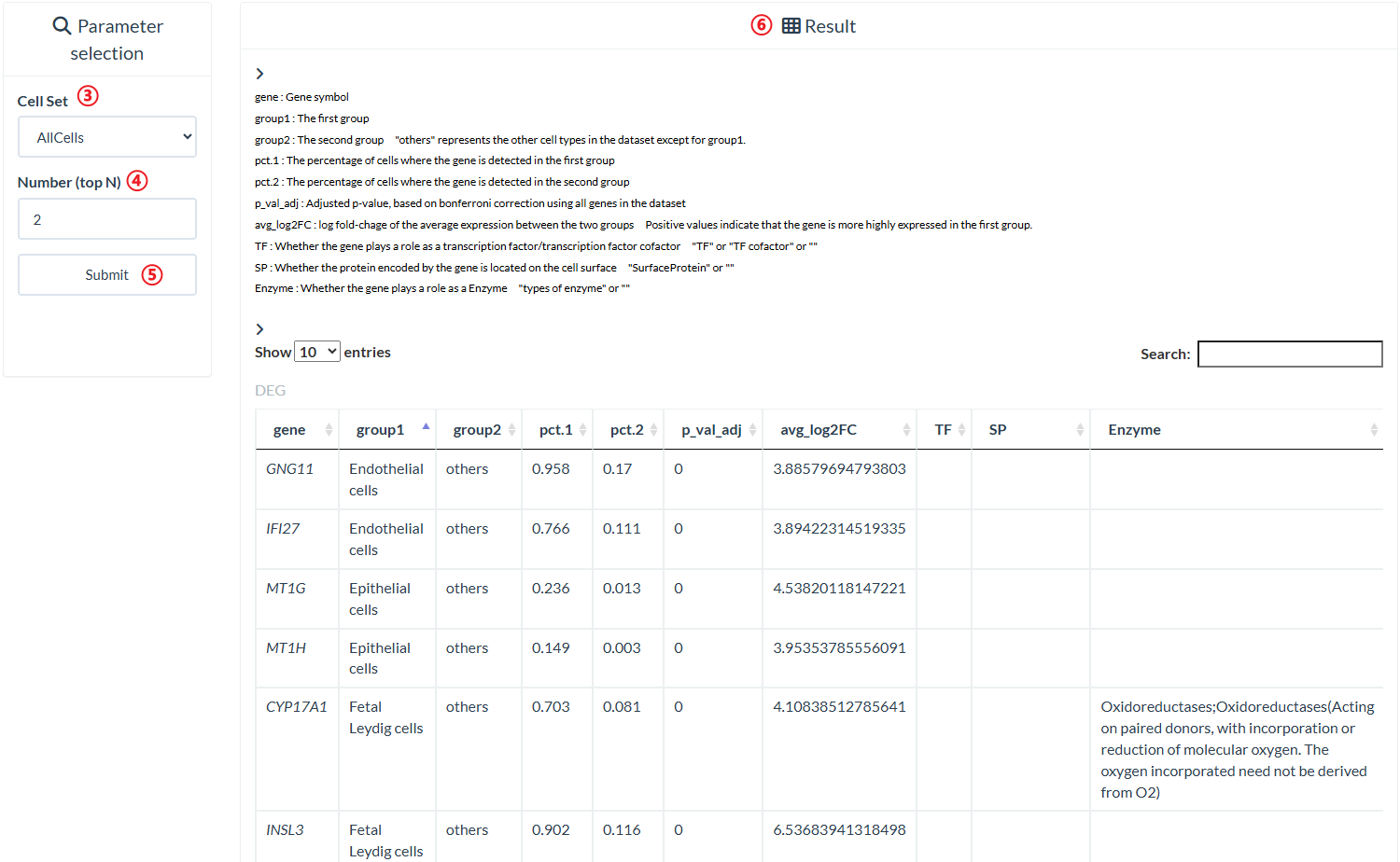
Users can query differentially expressed genes for each cell type in each cell set displayed on the "Datasets available" panel of the "Differentially Expressed Genes" page.
step1 : Click "Differentially Expressed Genes" in the pull-down menu of "TestisAtlas".
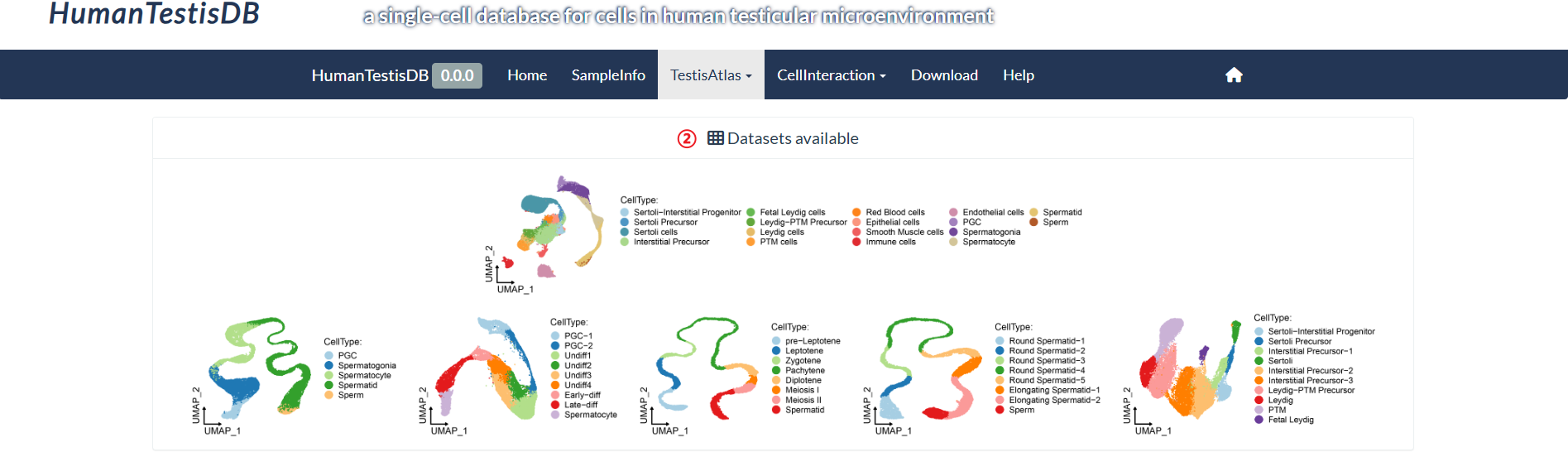
step2 : The "Datasets available" panel shows six cell sets with annotations.
step3 : Select the cell set to query in "Cell Set".
step4 : Input a number to query the top N differential gene of each cell group in "Number (top N)".
step5 : Click "Submit" to query result.
step6 : The result is on the right panel.
How to explore "Dynamically Expressed Genes" page?
Users can view or download genes dynamically expressed along different developmental trajectories, including:
Sertoli-Interstitial Progenitor-----Sertoli Precursor-----Sertoli
Sertoli-Interstitial Progenitor-----Interstitial Precursor-1
Interstitial Precursor-2-----Interstitial Precursor-3
Interstitial Precursor-2-----Fetal Leydig
Leydig-PTM Precursor-----Leydig
Leydig-PTM Precursor-----PTM
pre-Leptotene-----early Spermatocyte
Round Spermatid-1-----Sperm
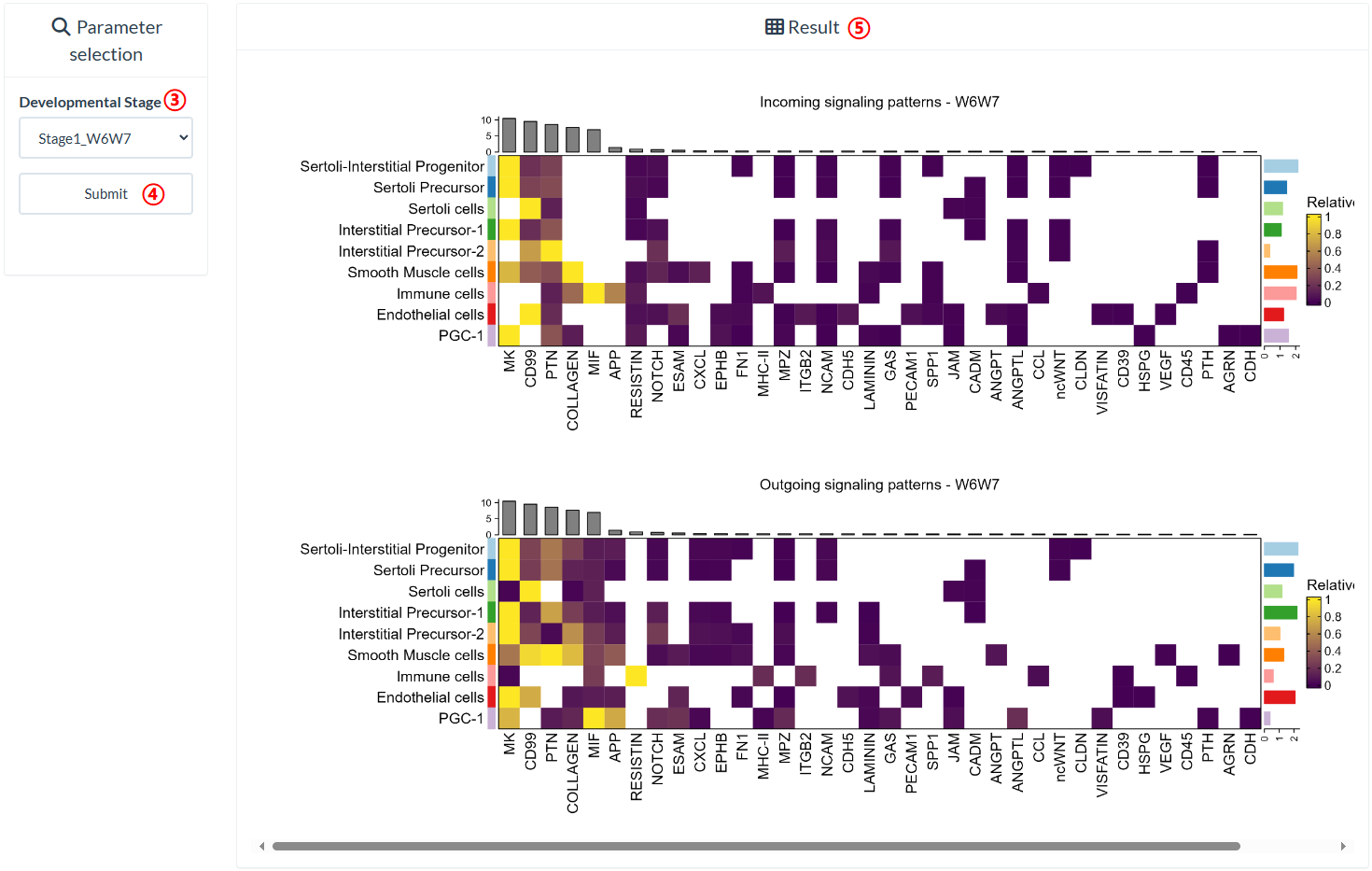
How to explore "Intra-stage Pathway" page?
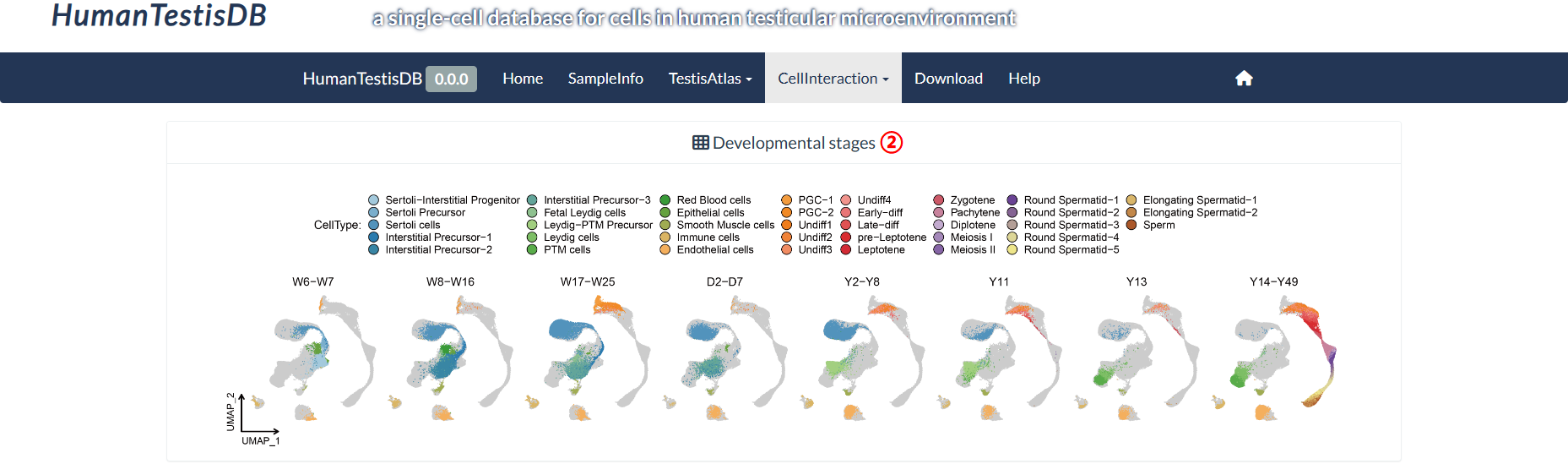
The top panel of the "Intra-stage Pathway" page shows the UMAP plots of eight distinct developmental stages that differ in their cell type composition. On the bottom panel, users can query the signals received or sent by each cell type within each developmental stage. The rows of the heatmap have been normalized, which means that the strengths of different signals received or sent by the same cell type are comparable.
step1 : Click "Intra-stage Pathway" in the pull-down menu of "CellInteraction".
step2 : The "Developmental stages" panel shows eight developmental stages that differ in cell type composition.
step3 : Select the developmental stage to query in "Developmental Stage".
step4 : Click "Submit" to query result.
step5 : The result is on the right panel.
How to explore "Inter-stage Pathway" page?
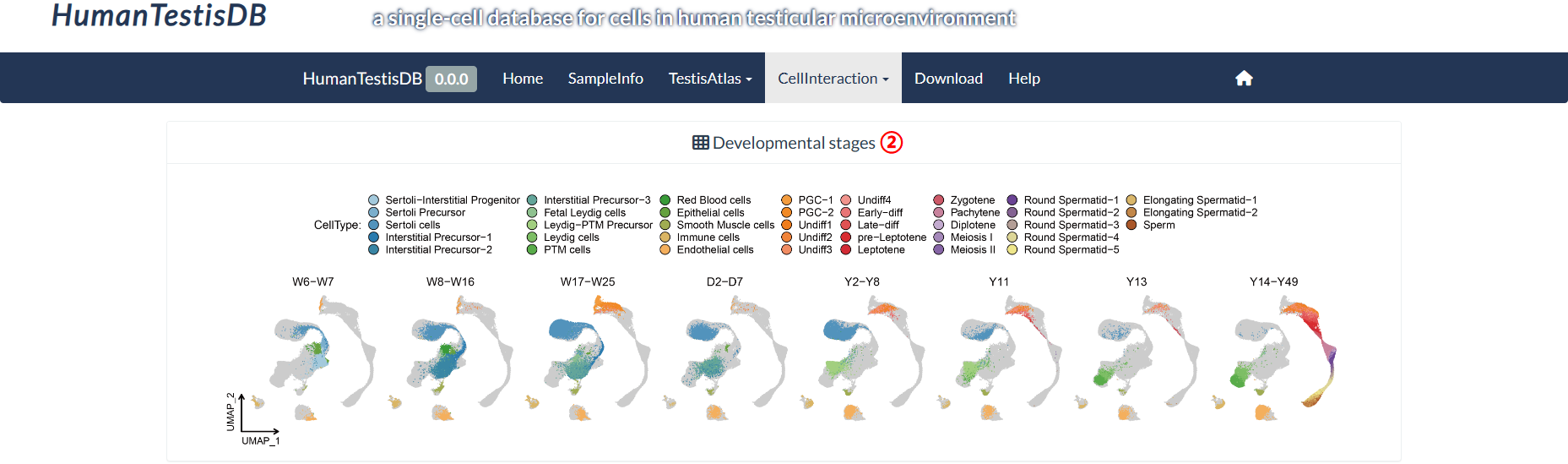
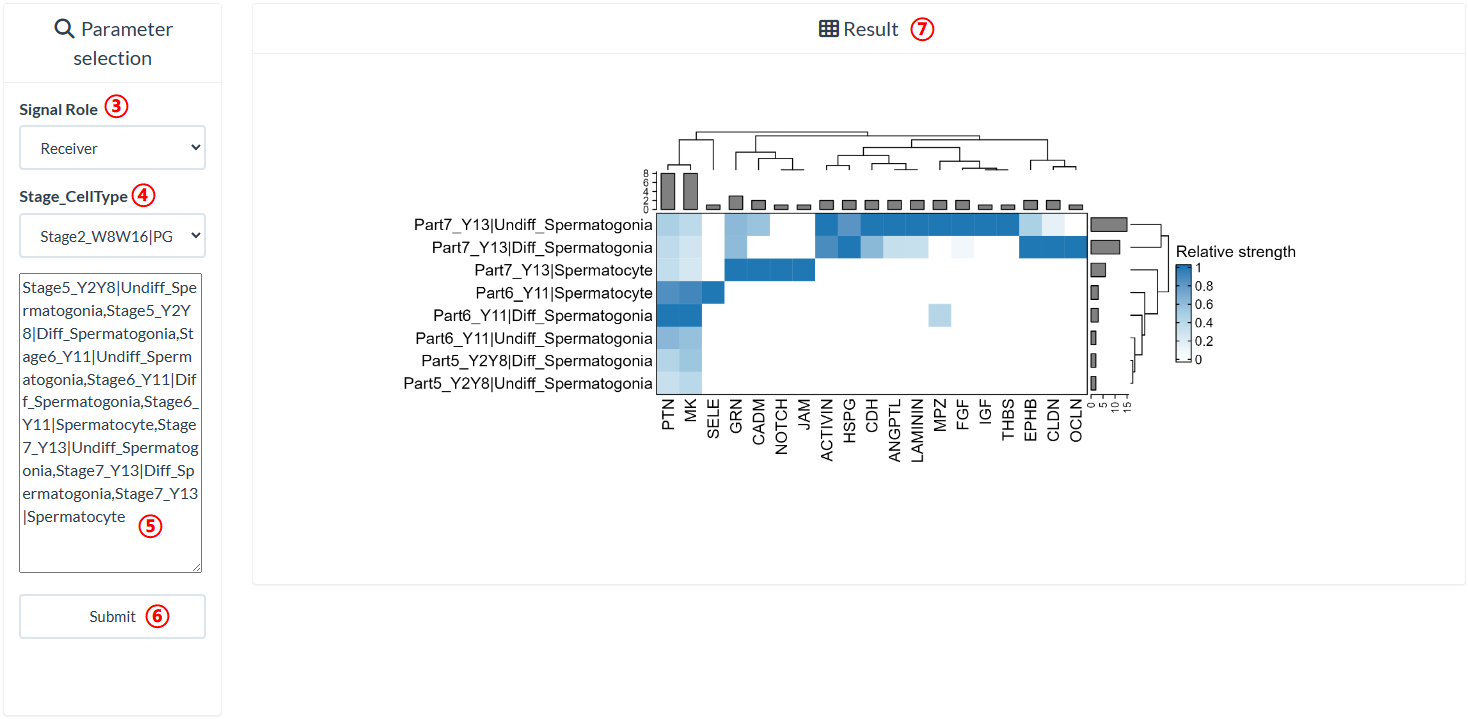
The top panel of the "Inter-stage Pathway" page shows the UMAP plots of eight distinct developmental stages that differ in their cell type composition. On the bottom panel, users can query the signals received or sent by different cell types at different developmental stages. The columns of the heatmap have been normalized, which means that the strengths of the same signal received or sent by different cell types are comparable. Note: If the value of "Stage_CellType" is empty, the right panel will display the results generated using the default value of "Stage5_Y2Y8|Undiff_Spermatogonia".
step1 : Click "Inter-stage Pathway" in the pull-down menu of "CellInteraction".
step2 : The "Developmental stages" panel shows eight developmental stages that differ in cell type composition.
step3 : Select a signal role of the cell type in "Signal Role".
step4-step5 : Select/Input one or more cell types existing at a developmental stage (using commas as the separator) in "Stage_CellType".
Provide two ways to select cell types. The first way is to select a cell type in the region marked as 4. The selected cell types will be displayed in the region marked as 5. The second way is to directly input the cell types to be queried in the region marked as 5, using commas as separators.
step6 : Click "Submit" to query result.
step7 : The result is on the right panel.
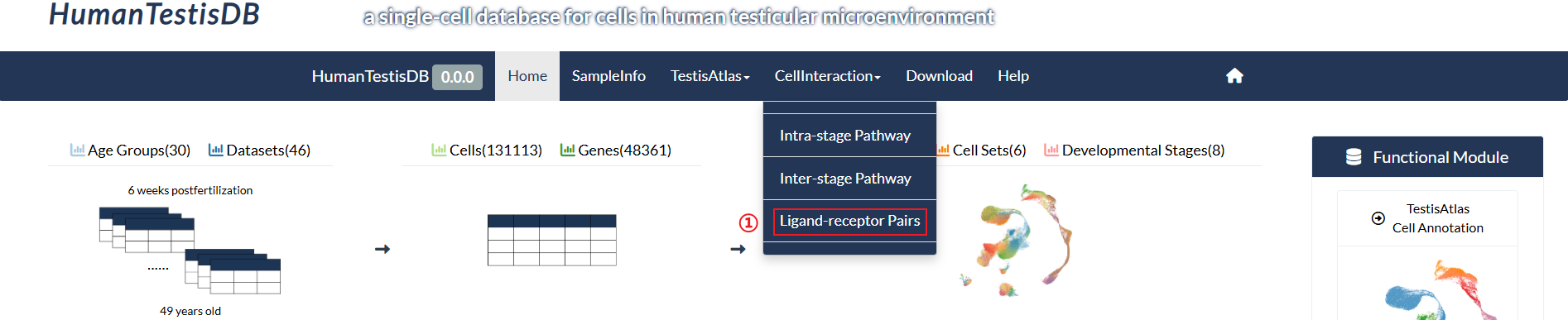
How to explore "Ligand-receptor Pairs" page?
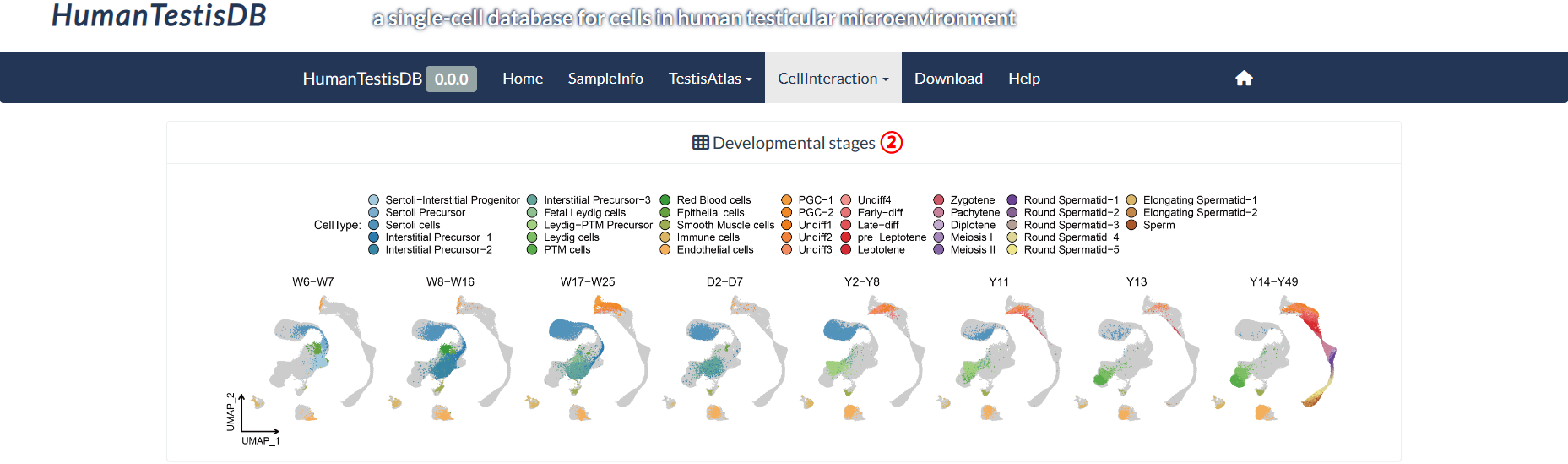
The top panel of the "Ligand-receptor Pairs" page shows the UMAP plots of eight distinct developmental stages that differ in their cell type composition. The bottom panel allows for detailed exploration of the signal pathways received or sent by each cell type within each developmental stage, such as receptor-ligand pairs and the cell types involved in these interactions. The values of the heatmap are not normalized, which are the communication probability calculated by the CellChat package.
step1 : Click "Ligand-receptor Pairs" in the pull-down menu of "CellInteraction".
step2 : The "Developmental stages" panel shows eight developmental stages that differ in cell type composition.
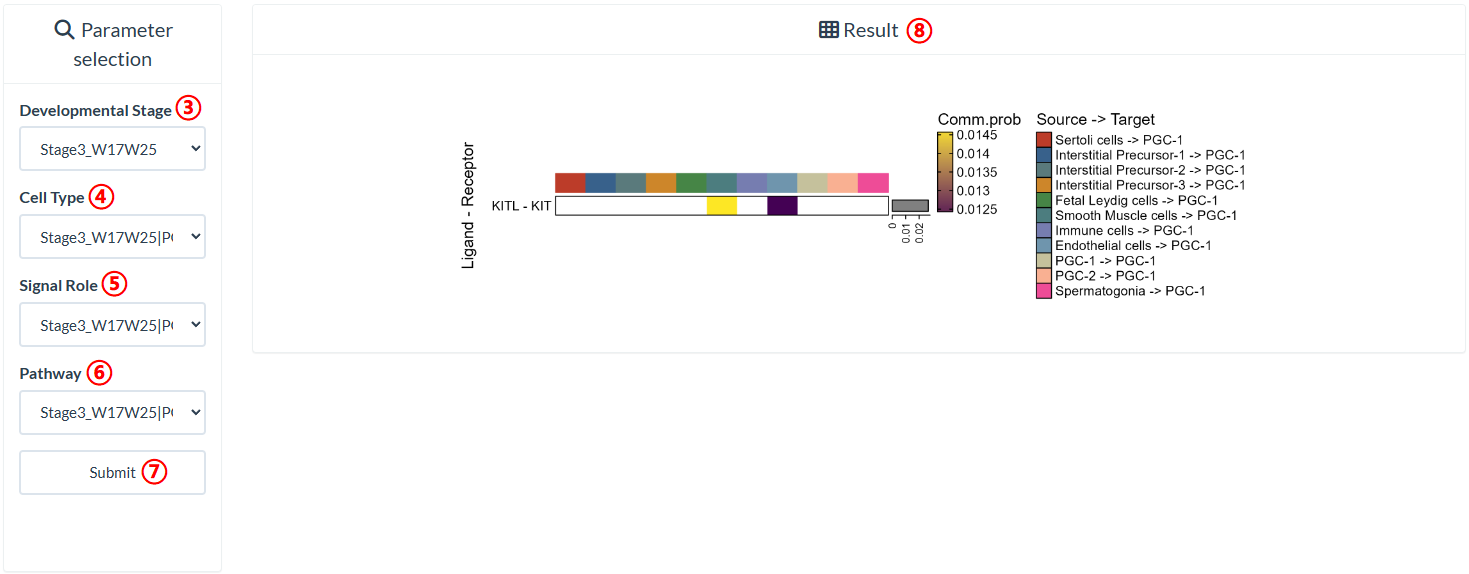
step3 : Select the developmental stage to query in "Developmental Stage".
step4 : Select a cell type existing in the developmental stage in "Cell Type".
step5 : Select a signal role of the cell type in "Signal Role".
step6 : Select a pathway related to the cell type in "Pathway".
step7 : Click "Submit" to query result.
step8 : The result is on the right panel.